From MIT, Scientists Preserve DNA in Amber-Like Polymer
For long-term storage
This is a Press Release edited by StorageNewsletter.com on June 18, 2024 at 2:01 pm![]() By Anne Trafton, MIT News
By Anne Trafton, MIT News
With their ‘T-REX’ (Thermoset-REinforced Xeropreservation) method, DNA embedded in the polymer could be used for long-term storage of genomes or digital data such as photos and music.
In the movie Jurassic Park, scientists extracted DNA that had been preserved in amber for millions of years, and used it to create a population of long-extinct dinosaurs.
Inspired partly by that film, MIT researchers have developed a glassy, amber-like polymer that can be used for long-term storage of DNA, whether entire human genomes or digital files such as photos.
Most current methods for storing DNA require freezing temperatures, so they consume a great deal of energy and are not feasible in many parts of the world. In contrast, the new amber-like polymer can store DNA at room temperature while protecting the molecules from damage caused by heat or water.
The researchers showed that they could use this polymer to store DNA sequences encoding the theme music from Jurassic Park, as well as an entire human genome. They also demonstrated that the DNA can be easily removed from the polymer without damaging it.
“Freezing DNA is the number one way to preserve it, but it’s very expensive, and it’s not scalable,” says James Banal, former postdoc, MIT. “I think our new preservation method is going to be a technology that may drive the future of storing digital information on DNA.”
Banal and Jeremiah Johnson, the A. Thomas Geurtin, professor of chemistry, MIT, are the senior authors of the study, published in the Journal of the American Chemical Society. Former MIT postdoc Elizabeth Prince and MIT postdoc Ho Fung Cheng are the lead authors of the paper.
Capturing DNA
DNA, a very stable molecule, is well-suited for storing massive amounts of information, including digital data. Digital storage systems encode text, photos, and other kind of information as a series of 0s and 1s. This same information can be encoded in DNA using the 4 nucleotides that make up the genetic code: A, T, G, and C. For example, G and C could be used to represent 0 while A and T represent 1.
DNA offers a way to store this digital information at very high density: In theory, a coffee mug full of DNA could store all of the world’s data. DNA is also very stable and relatively easy to synthesize and sequence.
In 2021, Banal and his postdoc advisor, Mark Bathe, an MIT professor of biological engineering, developed a way to store DNA in particles of silica, which could be labeled with tags that revealed the particles’ contents. That work led to a spinout called Cache DNA, Inc.
One downside to that storage system is that it takes several days to embed DNA into the silica particles. Furthermore, removing the DNA from the particles requires hydrofluoric acid, which can be hazardous to workers handling the DNA.
To come up with alternative storage materials, Banal began working with Johnson and members of his lab. Their idea was to use a type of polymer known as a degradable thermoset, which consists of polymers that form a solid when heated. The material also includes cleavable links that can be easily broken, allowing the polymer to be degraded in a controlled way.
“With these deconstructable thermosets, depending on what cleavable bonds we put into them, we can choose how we want to degrade them,” Johnson says.
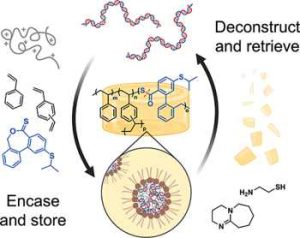
For this project, the researchers decided to make their thermoset polymer from styrene and a cross-linker, which together form an amber-like thermoset called cross-linked polystyrene. This thermoset is also very hydrophobic, so it can prevent moisture from getting in and damaging the DNA. To make the thermoset degradable, the styrene monomers and cross-linkers are copolymerized with monomers called thionolactones. These links can be broken by treating them with a molecule called cysteamine.
Because styrene is so hydrophobic, the researchers had to come up with a way to entice DNA – a hydrophilic, negatively charged molecule – into the styrene.
To do that, they identified a combination of three monomers that they could turn into polymers that dissolve DNA by helping it interact with styrene. Each of the monomers has different features that cooperate to get the DNA out of water and into the styrene. There, the DNA forms spherical complexes, with charged DNA in the center and hydrophobic groups forming an outer layer that interacts with styrene. When heated, this solution becomes a solid glass-like block, embedded with DNA complexes.
The researchers dubbed their method T-REX. The process of embedding DNA into the polymer network takes a few hours, but that could become shorter with further optimization, the researchers say.
To release the DNA, the researchers first add cysteamine, which cleaves the bonds holding the polystyrene thermoset together, breaking it into smaller pieces. Then, a detergent called SDS can be added to remove the DNA from polystyrene without damaging it.
Storing information
Using these polymers, the researchers showed that they could encapsulate DNA of varying length, from tens of nucleotides up to an entire human genome (more than 50,000 base pairs). They were able to store DNA encoding the Emancipation Proclamation and the MIT logo, in addition to the theme music from Jurassic Park.
After storing the DNA and then removing it, the researchers sequenced it and found that no errors had been introduced, which is a critical feature of any digital data storage system.
The researchers also showed that the thermoset polymer can protect DNA from temperatures up to 75°C (167°F). They are now working on ways to streamline the process of making the polymers and forming them into capsules for long-term storage.
Cache DNA, a company started by Banal and Bathe, with Johnson as a member of the scientific advisory board, is now working on further developing DNA storage technology. The earliest application they envision is storing genomes for personalized medicine, and they also anticipate that these stored genomes could undergo further analysis as better technology is developed in the future.
“The idea is, why don’t we preserve the master record of life forever?” Banal says. “Ten years or 20 years from now, when technology has advanced way more than we could ever imagine today, we could learn more and more things. We’re still in the very infancy of understanding the genome and how it relates to disease.”
The research was funded by the National Science Foundation.
Article: Reversible Nucleic Acid Storage in Deconstructable Glassy Polymer Networks
Journal of the American Chemical Society (JACS) has published an article written by Elisabeth Prince,Ho Fung Cheng, Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, United States, James L. Banal, Cache DNA, Inc., 733 Industrial Road, San Carlos, California 94070, United States, and Jeremiah A. Johnson, Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, United States.
Abstract: “The rapid decline in DNA sequencing costs has fueled the demand for nucleic acid collection to unravel genomic information, develop treatments for genetic diseases, and track emerging biological threats. Current approaches to maintaining these nucleic acid collections hinge on continuous electricity for maintaining low-temperature and intricate cold-chain logistics. Inspired by the millennia-long preservation of fossilized biological specimens in calcified minerals or glassy amber, we present Thermoset-REinforced Xeropreservation (T-REX): a method for storing DNA in deconstructable glassy polymer networks. Key to T-REX is the development of polyplexes for nucleic acid encapsulation, streamlining the transfer of DNA from aqueous to organic phases, replete with initiators, monomers, cross-linkers, and thionolactone-based cleavable comonomers required to form the polymer networks. This process successfully encapsulates DNA that spans different length scales, from tens of bases to gigabases, in a matter of hours compared to days with traditional silica-based encapsulation. Further, T-REX permits the extraction of DNA using comparatively benign reagents, unlike the hazardous hydrofluoric acid required for recovery from silica. T-REX provides a path toward low-cost, time-efficient, and long-term nucleic acid preservation for synthetic biology, genomics, and digital information storage, potentially overcoming traditional low-temperature storage challenges.“














 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter


