R&D: DNA Assembly for Nanopore Storage Readout
Assembly strategy is generalizable to any application that requires nanopore sequencing of small DNA amplicons.
This is a Press Release edited by StorageNewsletter.com on July 15, 2019 at 1:41 pmNature Communications has published an article written by Randolph Lopez, Department of Bioengineering, University of Washington, Seattle, WA, 98105, USA, and Molecular Engineering and Sciences Institute, University of Washington, Seattle, WA, 98195, USA, Yuan-Jyue Chen, Siena Dumas Ang, Sergey Yekhanin, Konstantin Makarychev, Miklos Z Racz, Microsoft Research, Redmond, WA, 98052, USA, Georg Seelig, Molecular Engineering and Sciences Institute, University of Washington, Seattle, WA, 98195, USA, Department of Electrical and Computer Engineering, University of Washington, Seattle, WA, 98195, USA, and Paul G. Allen School for Computer Science and Engineering, University of Washington, Seattle, WA, 98195, USA, Karin Strauss, Microsoft Research, Redmond, WA, 98052, USA, and Luis Ceze, Paul G. Allen School for Computer Science and Engineering, University of Washington, Seattle, WA, 98195, USA
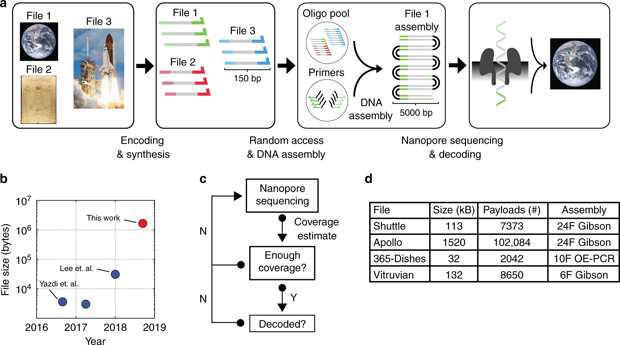
Overview of the DNA data storage workflow. a The encoding process starts with mapping multiple digital files into 150-nucleotide DNA sequences and sending them for synthesis. Each file has unique sequence addresses at the 5′ and 3′ end of each oligonucleotide for random access retrieval. Using PCR primers containing complementary overhang sequences, a specific file can be amplified and concatenated into long double-stranded DNA molecules suited for ONT Nanopore sequencing. Upon sequencing, a subset of reads with high accuracy are used to decode the selected file. b Our assembly and decoding strategy enabled successful decoding of 1.67MB of digital information stored in DNA using nanopore sequencing. Our work represents a 2-order of magnitude improvement in demonstrated decoding ability using nanopore sequencing for DNA storage. c Sequence-until diagram. Nanopore sequencing enables real-time coverage estimation for decoding of digital files store in DNA. This enables the user to generate reads until coverage is enough for successful decoding. Upon decoding, a different file can be sequenced in the same flowcell or the sequencing run can be stopped and resumed later on. d Four different files encoded in DNA were amplified, assembled and sequenced using ONT MinION platform. We implemented overlap-extension PCR and Gibson Assembly to build assemblies of 6, 10, or 24 fragments for each file
Abstract: “Synthetic DNA is becoming an attractive substrate for digital data storage due to its density, durability, and relevance in biological research. A major challenge in making DNA data storage a reality is that reading DNA back into data using sequencing by synthesis remains a laborious, slow and expensive process. Here, we demonstrate successful decoding of 1.67 megabytes of information stored in short fragments of synthetic DNA using a portable nanopore sequencing platform. We design and validate an assembly strategy for DNA storage that drastically increases the throughput of nanopore sequencing. Importantly, this assembly strategy is generalizable to any application that requires nanopore sequencing of small DNA amplicons.“













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

