R&D: World First for Reading Digitally Encoded Synthetic Molecules
Paves way for synthetic polymer storage of several kilobytes of data, roughly equivalent to page of text-just like first floppy disks.
This is a Press Release edited by StorageNewsletter.com on October 26, 2017 at 2:46 pmFrom CNRS (Centre National de la Recherche Scientifique)
For the first time, using mass spectrometry, researchers have successfully read several bytes (1) of data recorded on a molecular scale using synthetic polymers.
By inserting fragile bonds between each molecular ‘byte,’
digital polymers may be easily read through mass spectrometry.
© Jean-François Lutz, Institut Charles Sadron (CNRS)
Their work, conducted under the aegis of the Institut Charles Sadron (CNRS) in Strasbourg, France and the Institute of Radical Chemistry (CNRS/Aix-Marseille University, France), sets a benchmark for the amount of data – stored as a sequence of molecular units (monomers)—that may be read using this routine method. It also sets the stage for data storage on a scale 100 times smaller than that of current hard drives. The scientists’ findings are published in Nature Communications on October 17, 2017 (see below).
Will hard drives one day use polymers for data storage? For several years now, researchers have faced the challenge of trying to record information with these long molecules. Polymers have great potential since, to record a bit, (1) their component monomers require 100 times less space than current HDDs. Their use could therefore reduce the size of computer storage media. Yet researchers have been thwarted in their efforts to effectively read polymer data. Now a team of CNRS and Aix-Marseille University scientists has demonstrated that a mass spectrometer may be used to read long sequences of data recorded on a molecular scale. This is a breakthrough because mass spectrometers, popular among chemists, are fast and easy to use.
To pull it off, the team used synthetic molecules, simpler to work with than natural molecules like DNA. Their structure was optimized for sequencing by mass spectrometry. The polymers are made up of two kinds of monomers (with phosphate groups)—corresponding to 0 and 1 respectively. After every eight of these monomer ‘bits,’ a molecular separator was added. The number of bytes represented by the complete polymer equals the number of eight-bit groups. The first step in reading the encoded information is to divide the polymer into molecular bytes by snapping it apart at the separator sites; the next is to break the phosphate bonds, for sequencing of each byte.
The team of chemists managed to synthesize polymers that can store up to eight bytes. Thus, they were able to record the word ‘Sequence’ in ASCII code, which assigns a unique byte to each letter and punctuation mark. By successfully decoding this word using mass spectrometry, they set a new record for the length of a molecule that may be read using this technique. Although manual analysis of the digital data does take a few hours, it should be possible to reduce the time needed to a few milliseconds by developing software to perform this task. By associating short read times with current automated methods for writing data, this work paves the way for synthetic polymer storage of several kilobytes of data, roughly equivalent to a page of text – just like the very first floppy disks.
(1 )A byte is a segment of information consisting of eight subunits called bits. Bits, which represent the lowest level of data, can each have only one of two possible values: 0 or 1.
Article: Mass spectrometry sequencing of long digital polymers
facilitated by programmed inter-byte fragmentation
Nature Communications as published an article written by Abdelaziz Al Ouahabi, Université de Strasbourg, CNRS, Institut Charles Sadron UPR22, 23 rue du Loess, 67034, Strasbourg, France , Jean-Arthur Amalian, Laurence Charles, Aix-Marseille Université, CNRS, UMR 7273, Institute of Radical Chemistry, 13397, Marseille, France, and Jean-François Lutz, Université de Strasbourg, CNRS, Institut Charles Sadron UPR22, 23 rue du Loess, 67034, Strasbourg, France.
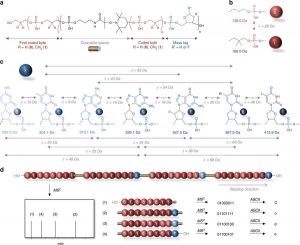
General concept studied herein for the sequencing of long digital polymer chains. a Molecular structure of the sequence-coded polymers prepared by automated phosphoramidite chemistry. These digital polymers contain n + 1 coded bytes noted in red. A byte is a sequence of eight coded monomers that represent 8 bits. Two consecutive bytes are separated by a linker noted in black, which contains a NO-C bond that can be preferentially cleaved during MS/MS analysis. In order to sort out the bytes after MS/MS cleavage, n bytes of the sequence are labeled with a mass tag noted in blue. b Molecular structure and mass of the two coded synthons that define the binary code in the polymers. c Molecular structure and mass of the mass tags that are used as bytes labels. In order to induce identifiable mass shifts after MS/MS cleavage, the mass of a byte tag (noted in blue) shall not be a multiple of 28, which is the mass difference between a 0 and a 1 coded unit. In addition, the mass difference between two tags (noted in grey) shall not be a multiple of 28. d Schematic representation of the mass spectrometry sequencing of a digital polymer containing 4 bytes of information. The polymer is first analyzed in MS/MS conditions, which lead to the favored cleavage of the weak NO-C bonds (depicted in yellow inside the grey spacers). Since they carry mass tags, the resulting cleaved bytes are sorted out by mass in the MS/MS spectrum (the displayed MS2 cartoon is idealized for clarity). Afterwards, each byte can be easily sequenced in MS3 conditions and the whole binary sequence can be deciphere.
Click to enlarge
Abstract: “In the context of data storage miniaturization, it was recently shown that digital information can be stored in the monomer sequences of non-natural macromolecules. However, the sequencing of such digital polymers is currently limited to short chains. Here, we report that intact multi-byte digital polymers can be sequenced in a moderate resolution mass spectrometer and that full sequence coverage can be attained without requiring pre-analysis digestion or the help of sequence databases. In order to do so, the polymers are designed to undergo controlled fragmentations in collision-induced dissociation conditions. Each byte of the sequence is labeled by an identification tag and a weak alkoxyamine group is placed between two bytes. As a consequence of this design, the NO-C bonds break first upon collisional activation, thus leading to a pattern of mass tag-shifted intact bytes. Afterwards, each byte is individually sequenced in pseudo-MS3 conditions and the whole sequence is found.“









 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

